Phylogenomic analysis restructures the Ulvophyceae
Gulbrandsen, Ø., Andresen, I. J., Krabberød, A. K., Bråte, J. and Shalchian-Tabrizi, K. 2021. Journal of Phycology. doi:10.1111/jpy.13168k
Abstract
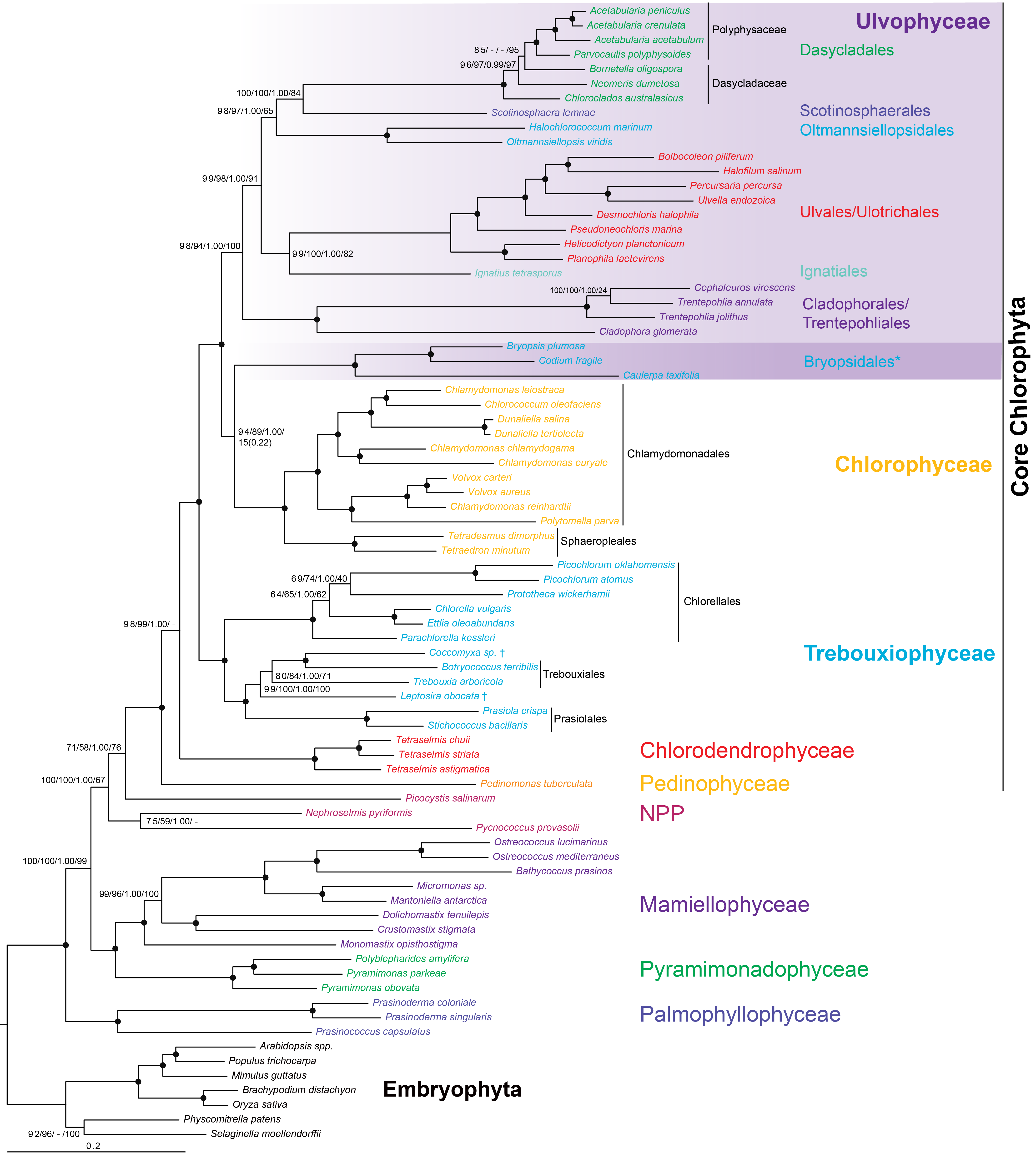
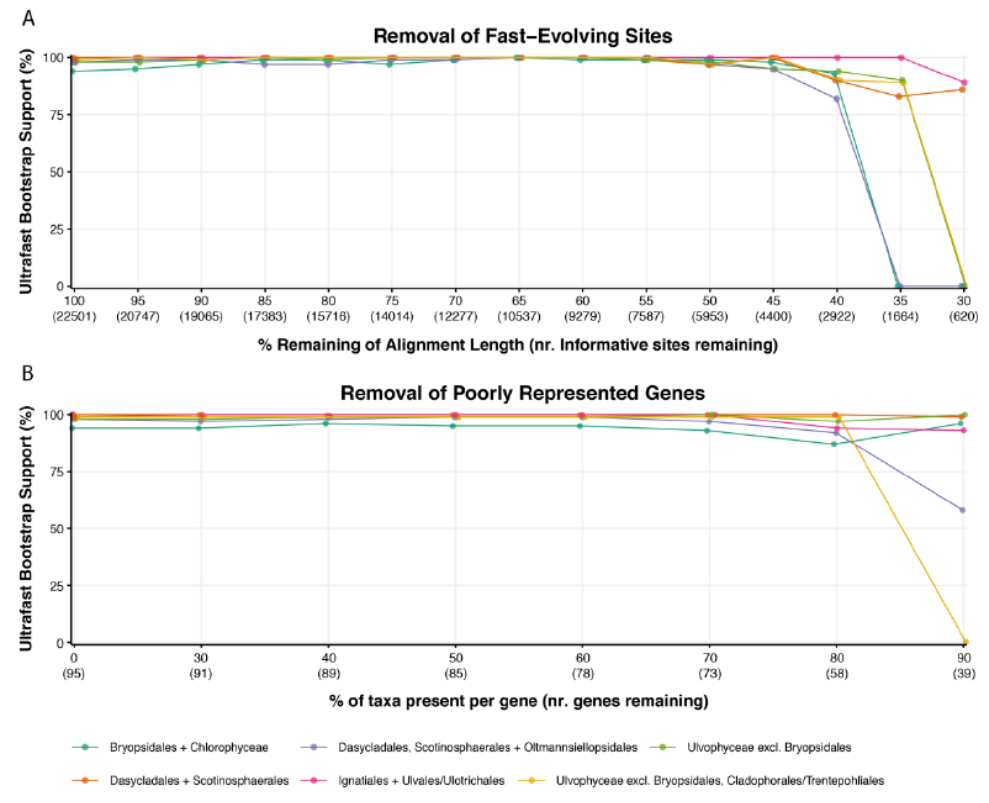
Here we present new transcriptome sequencing data from seven species of Dasycladales (Ulvophyceae) and a phylogenomic analysis of the Chlorophyta with a particular focus on Ulvophyceae. We have focused on a broad selection of green algal groups and carefully selected genes suitable for reconstructing deep eukaryote evolutionary histories. Increasing the taxon sampling of Dasycladales restructures the Ulvophyceae by identifying Dasycladales as closely related to Scotinosphaerales and Oltmannsiellopsidales. Contrary to previous studies, we do not find support for a close relationship between Dasycladales and a clade of Cladophorales and Trentepohliales. Instead, these groups are sister to the remainder of the Ulvophyceae. Furthermore, our analyses show high and consistent statistical support for a sister relationship between Bryopsidales and Chlorophyceae in trees generated with both homogeneous and heterogeneous (heterotachy) evolutionary models. Our study provides a new framework for a macroevolutionary interpretation of the evolutionary history of Ulvophyceae and the evolution of macroscopic cellular morphologies.