Genome sequencing and de novo assembly of the giant unicellular alga Acetabularia acetabulum using droplet MDA
Andresen, I. J., Orr. R. J. S., Krabberød, A. K., Shalchian-Tabrizi, K. and Bråte, J. 2021. Scientific Reports. doi:10.1038/s41598-021-92092-4
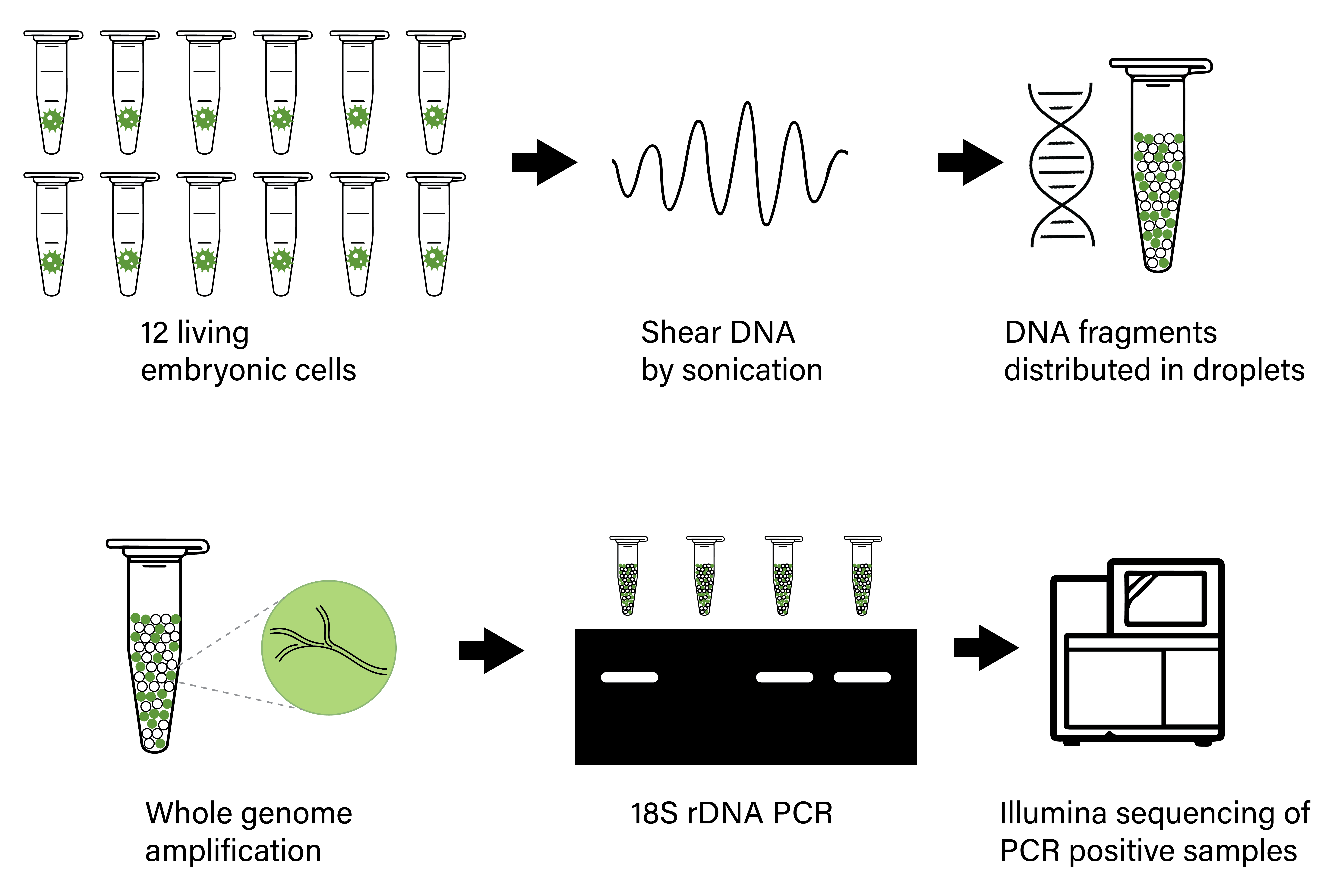
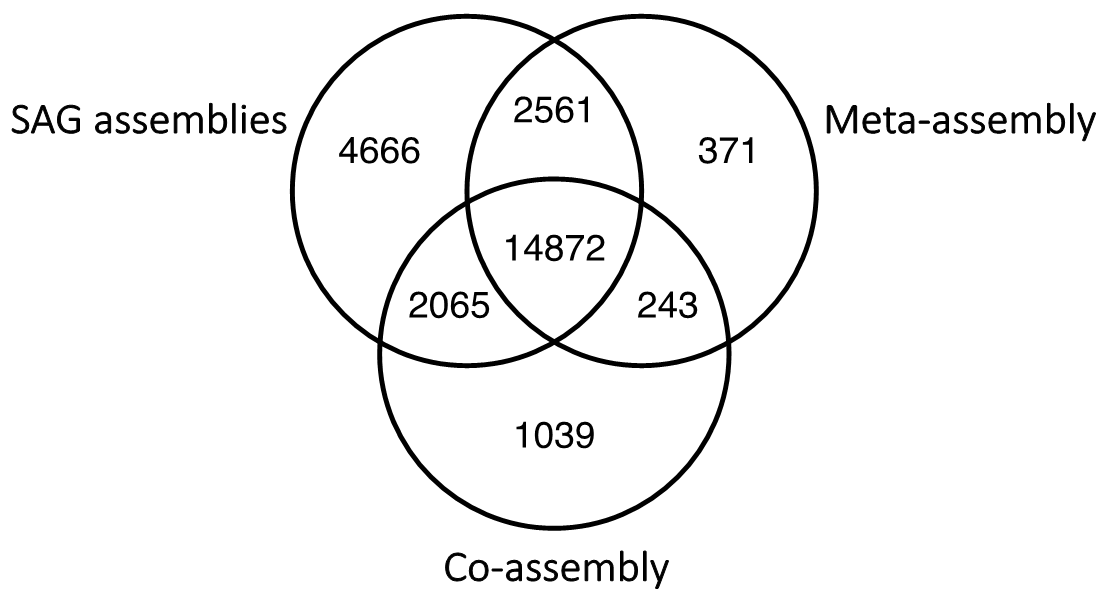
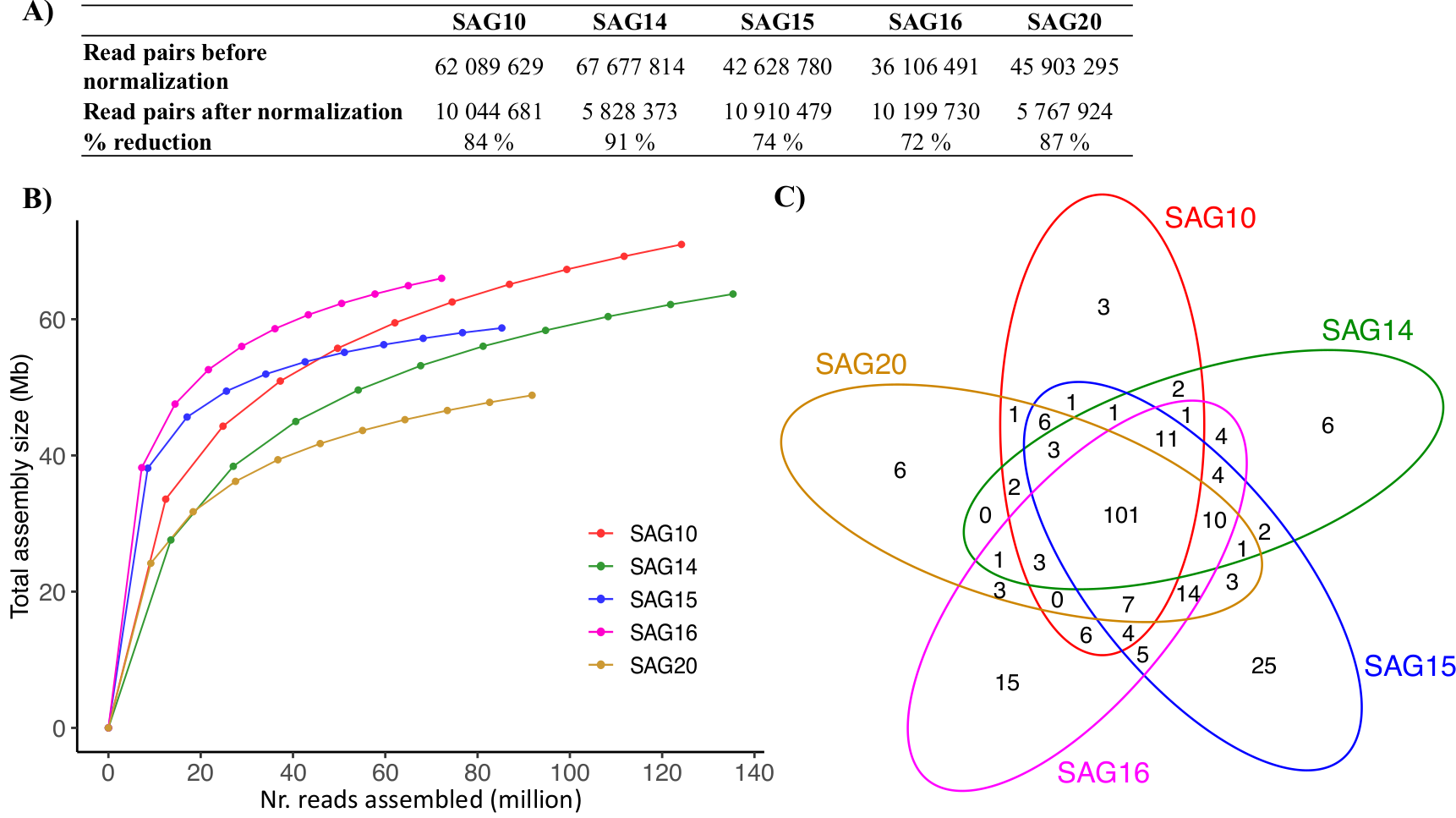
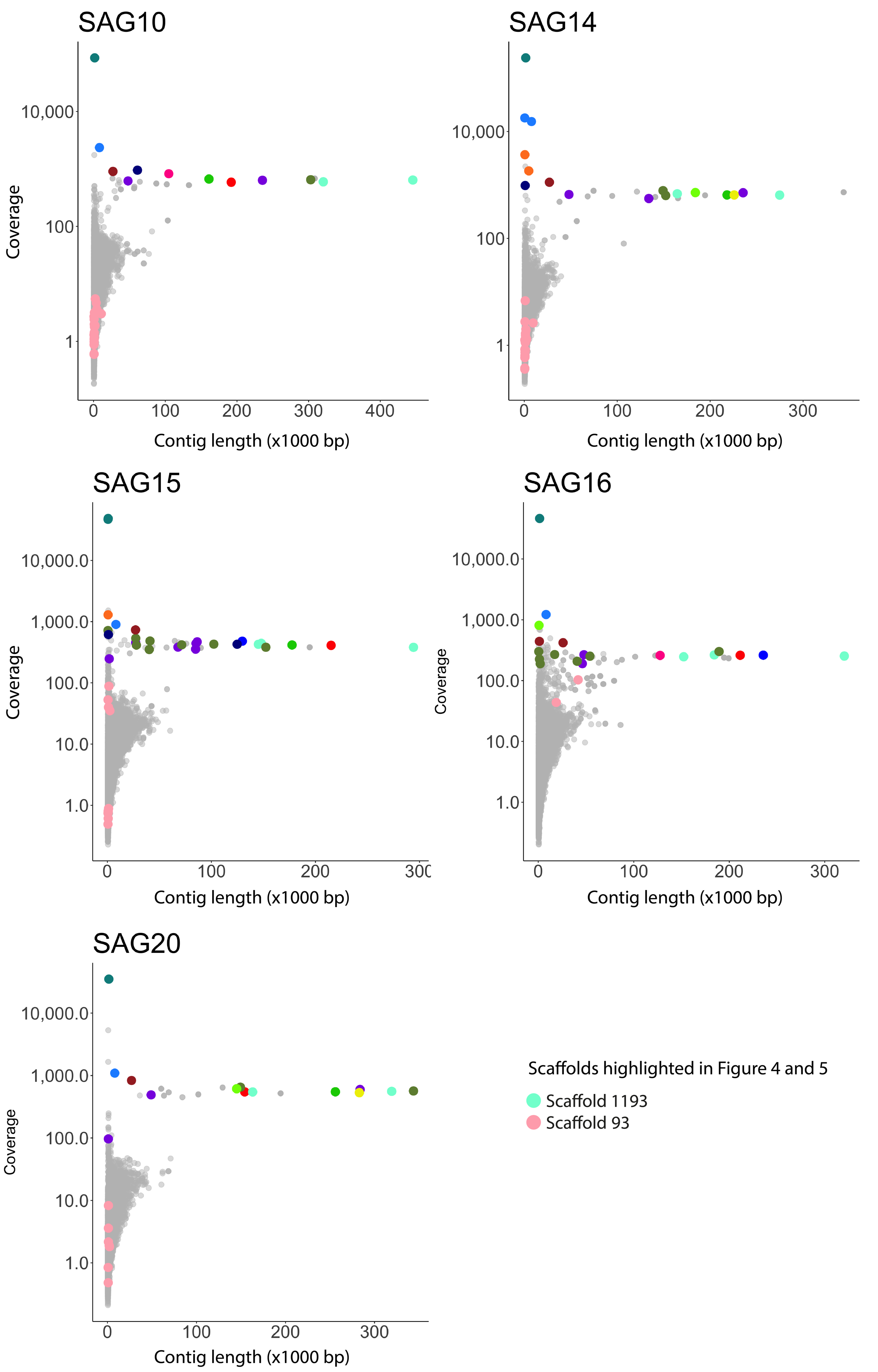
Abstract
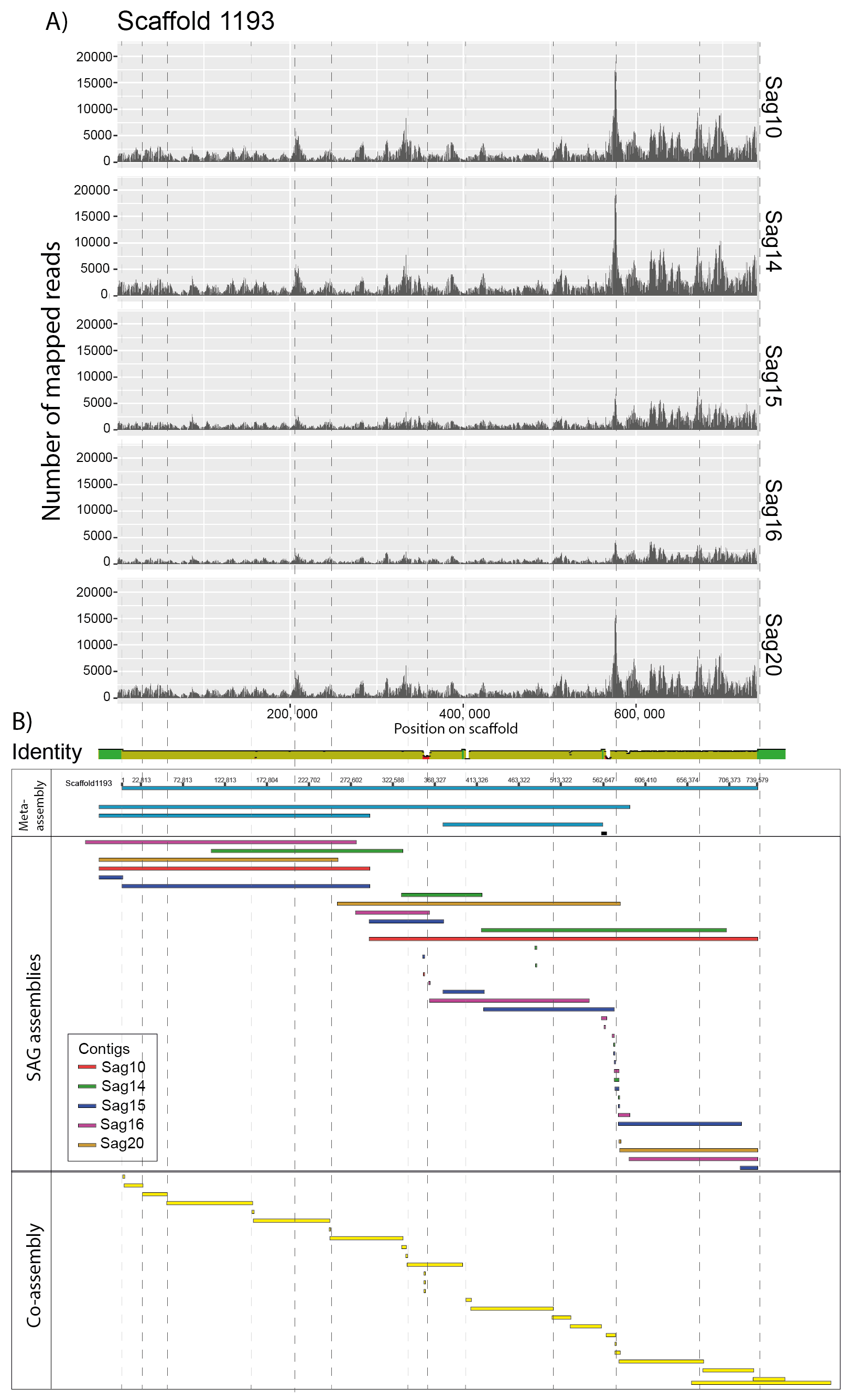
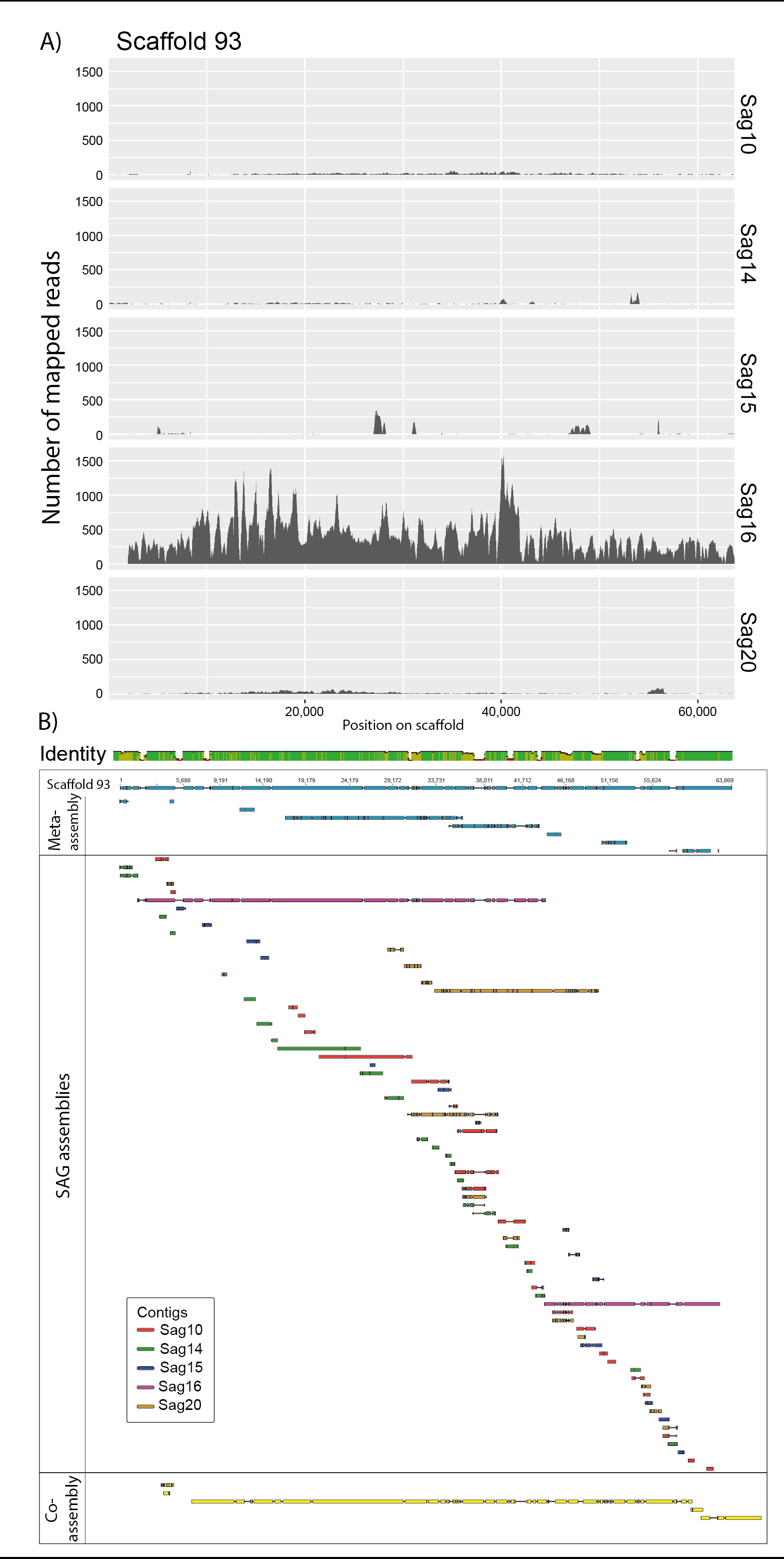
The macroscopic single-celled green alga Acetabularia acetabulum has been a model system in cell biology for more than a century. However, no genomic information is available from this species. Since the alga has a long life cycle, is difficult to grow in dense cultures, and has an estimated diploid genome size of almost 2 Gb, obtaining sufficient genomic material for genome sequencing is challenging. Here, we have attempted to overcome these challenges by amplifying genomic DNA using multiple displacement amplification (MDA) combined with microfluidics technology to distribute the amplification reactions across thousands of microscopic droplets. By amplifying and sequencing DNA from five single cells we were able to recover an estimated ~ 7–11% of the total genome, providing the first draft of the A. acetabulum genome. We highlight challenges associated with genome recovery and assembly of MDA data due to biases arising during genome amplification, and hope that our study can serve as a reference for future attempts on sequencing the genome from non-model eukaryotes.