Research
Updated 2025.08.29
Current projects
A lot of my time I spend developing tools for genomic analyses of blood borne viruses such as Hepatitis C Virus. And currently I am developing a Nextflow pipeline for assembly, genotyping and antiviral resistance analysis of HCV called HCVTyper. I am also doing research on the evolution and dispersal of HCV in Norway.
Previous projects
SARS-CoV-2
I was hired at the Norwegian Institute of Public Health during the Covid-19 pandemic and most of my time was spend tackling the vast amount of data that we produced from genome sequencing of SARS-CoV-2 in Norway. But I was also involved in a few research projects on SARS-CoV-2, such as our study on the dynamics and evolution of the UK-lineage in Norway, or this on the dynamics of the Norwegian AY.63 lineage, or one study where we discovered a novel Delta/Omicron recombinant.
Regulatory RNAs and the origin of animals
As a post doc I led two fully funded projects on genome regulation and the evolutionary origin of animals. We studied questions such as, How did the first animal look like, and how did it evolve from single-celled species? In particular. we were interested in the genes that regulate how other genes are used and how these may have contributed to the evolution of increased morphological and developmental complexity. We worked on sponges, comb jellies and the single-celled choanozoans, the closest ancestors of animals.
MicroRNAs
MicroRNAs are short RNA molecules that regulate other genes, and in animals microRNAs are an essential part of the genetic toolkit. I am trying to find out when microRNAs and the complementary protein machinery evolved in animals. We have recently discovered for the first time microRNAs, as well as homologs of the animal protein machinery which processes the miRNAs, in unicellular ancestors of animals, suggesting that these evolved much earlier than previously thought. The paper has been published in Current Biology.

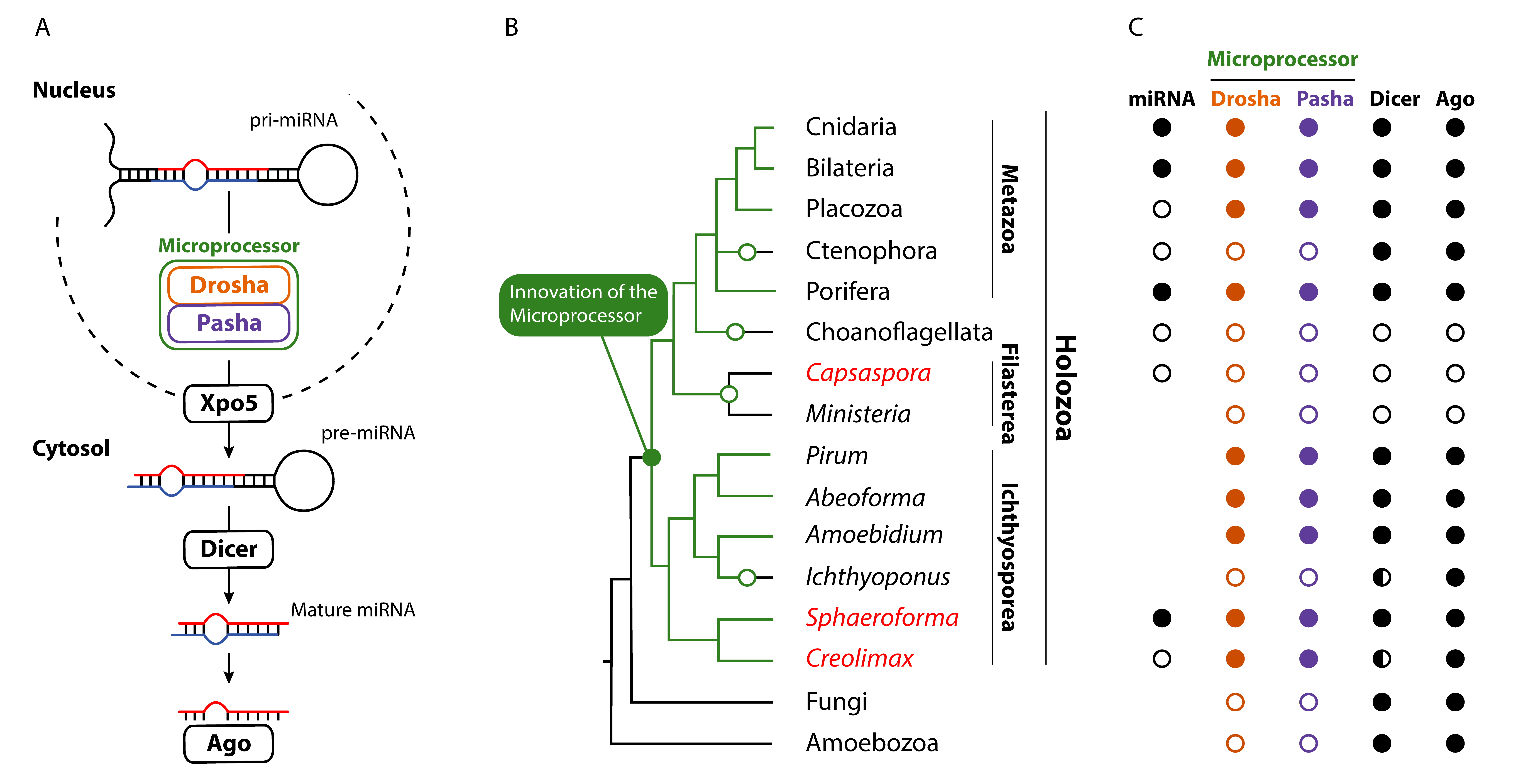
And we have also extended these results to show that microRNAs are in fact present across Ichthyosporea (currently only available as a preprint).
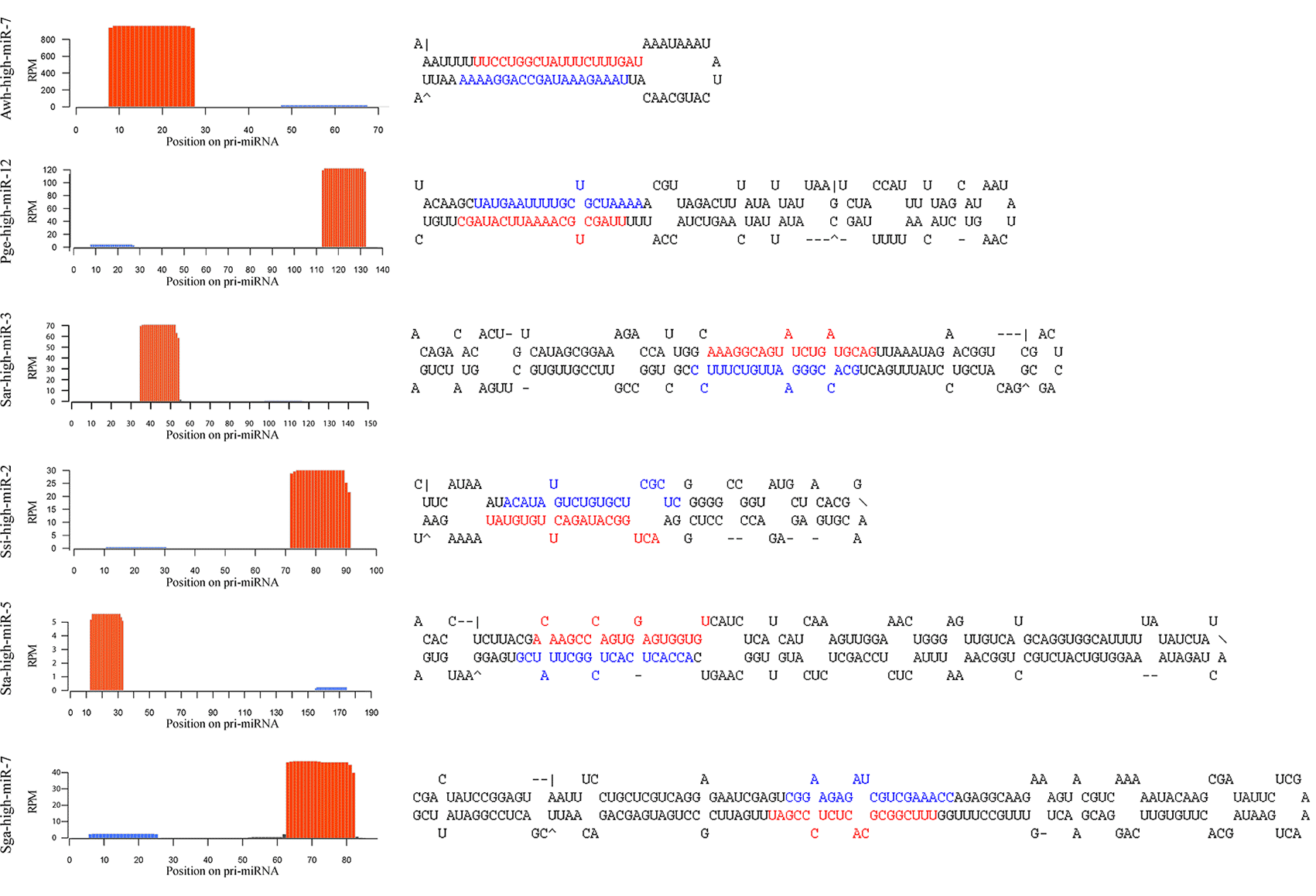
Long non-coding RNAs
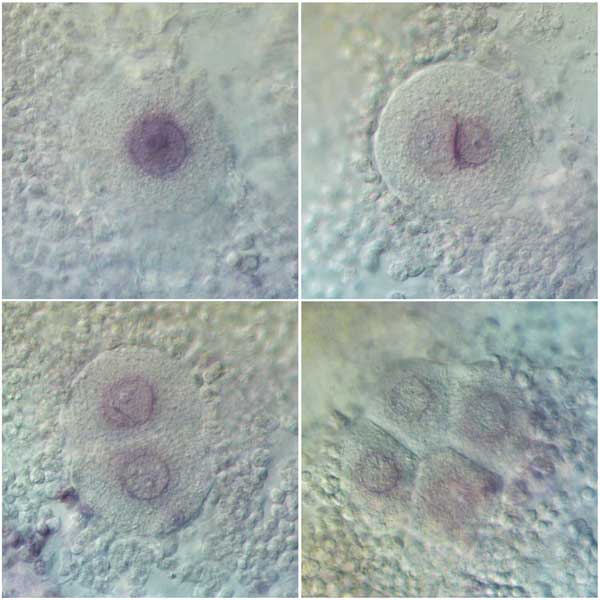
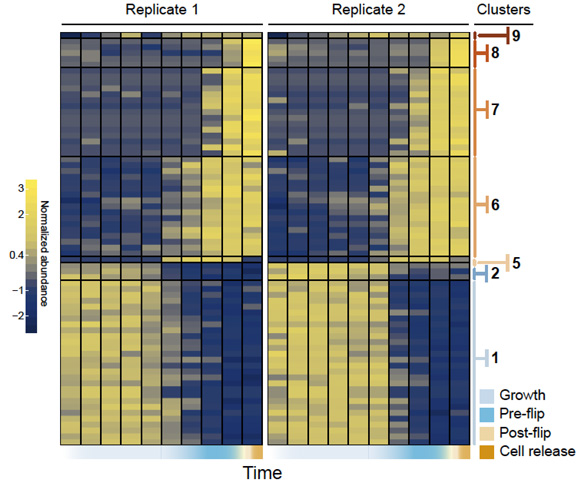
Another type of regulatory genes are long non-coding RNAs. This is a class of recently discovered RNA genes. Very little is known about what these genes do, and even less is known about their evolutionary significance. To get a glimpse of how the last common ancestor of animals might have used lncRNAs, we have studied their expression dynamics during cellular development in the sponge Sycon ciliatum (published in Proceedings B) and in the ichthyosporean Sphaeroforma arctica (published in eLife).
Protist diverst and evolution
I did my PhD on the diversity and evolution of protists, single-celled eukaryotes. In particular, I studied weird groups of protists that were largely unknown. I used methods such as environmental sequencing and molecular phylogenetics.