The impact of global lineage dynamics, border restrictions, and emergence of the B.1.1.7 lineage on the SARS-CoV-2 epidemic in Norway
Osnes, N. M., Alfsnes, K. Bråte, J., Garcia, I., Riis, R. K., Instefjord, K. H., Elshaug, H., Vollan, H. S., Moen, L. V., Pedersen, B. N., Caugant, D. A., Stene-Johansen, K., Hungnes, O., Bragstad, K., Brynildsrud, O. B. and Eldholm, V. 2021. Virus Evolution. doi:10.1093/ve/veab086
Abstract
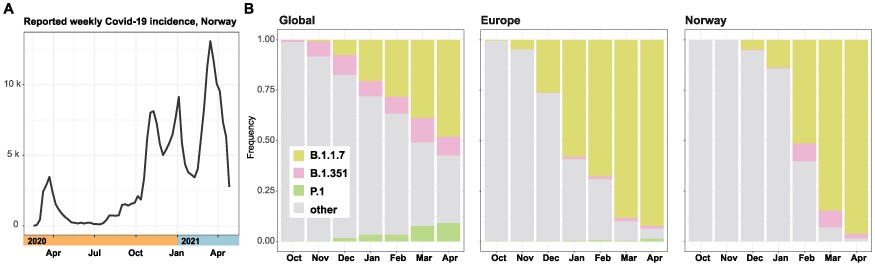
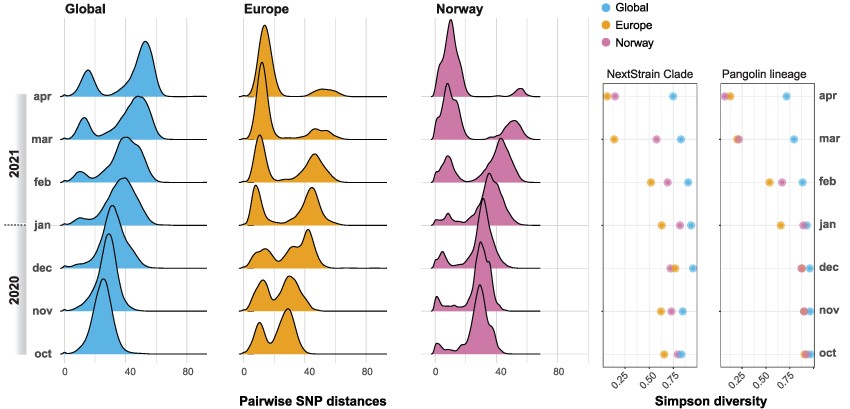
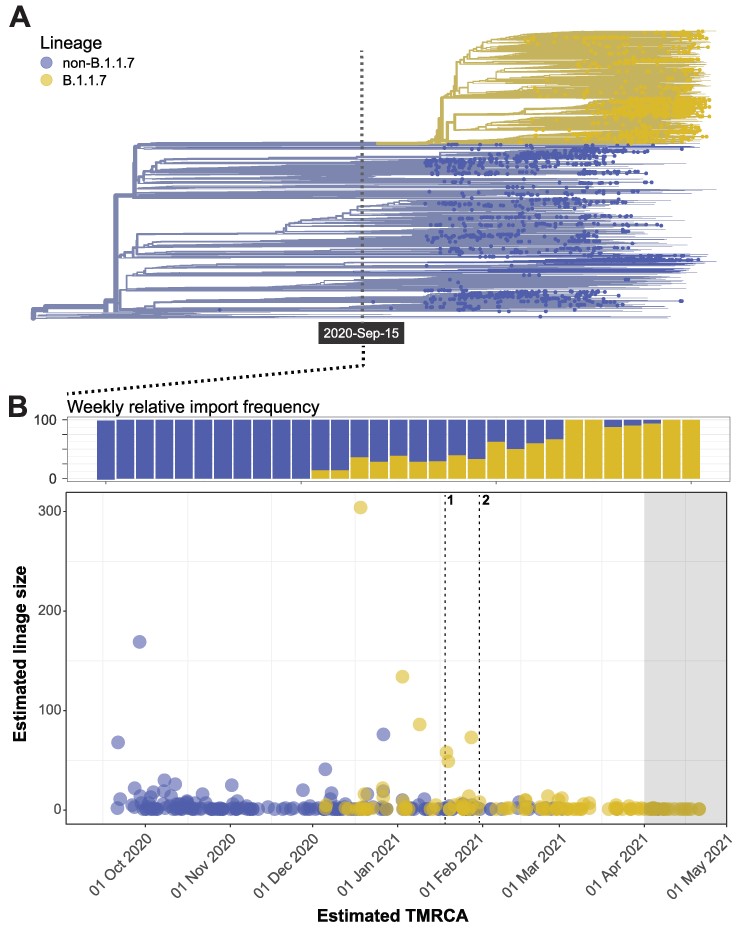
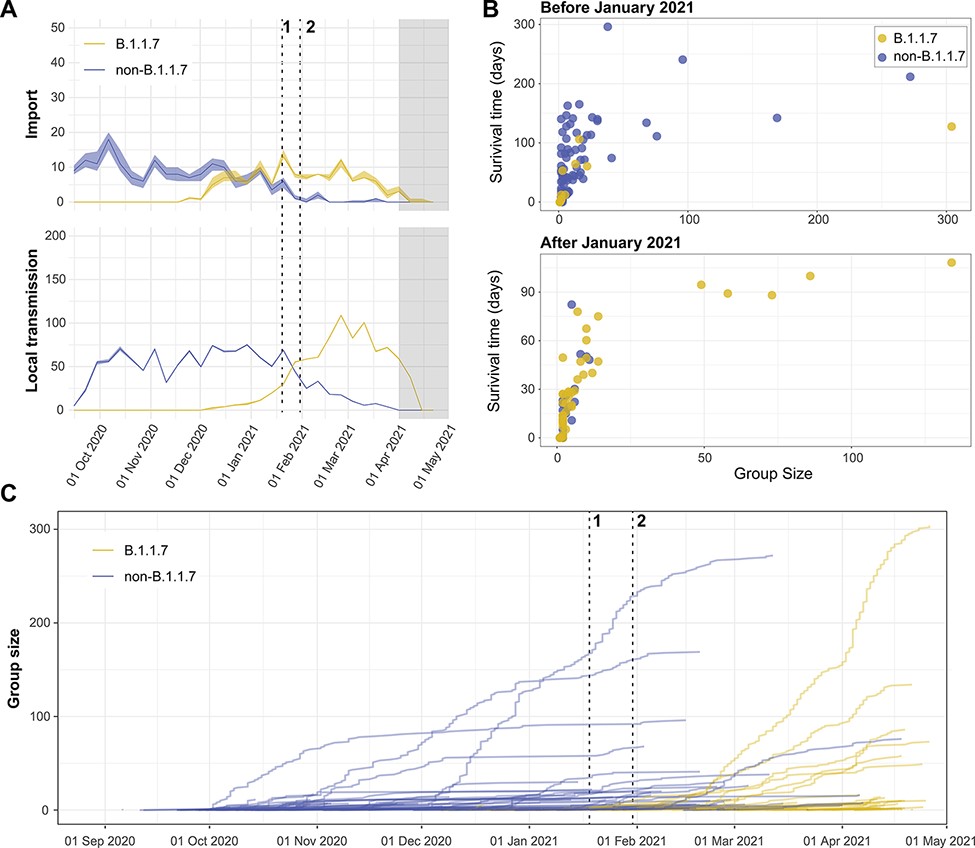
As the COVID-19 pandemic swept through an immunologically naïve human population, academics and public health professionals scrambled to establish methods and platforms for genomic surveillance and data sharing. This offered a rare opportunity to study the ecology and evolution of SARS-CoV-2 over the course of the ongoing pandemic. Here, we use population genetic and phylogenetic methodology to characterize the population dynamics of SARS-CoV-2 and reconstruct patterns of virus introductions and local transmission in Norway against this backdrop. The analyses demonstrated that the epidemic in Norway was largely import driven and characterized by the repeated introduction, establishment, and suppression of new transmission lineages. This pattern changed with the arrival of the B.1.1.7 lineage, which was able to establish a stable presence concomitant with the imposition of severe border restrictions.