Single cell transcriptomics, mega-phylogeny and the genetic basis of morphological innovations in Rhizaria
Krabberød, A. K., Orr, R. J. S, Bråte, J., Kristensen, T., Bjørklund, K. R and Shalchian-Tabrizi, K. 2017. Molecular Biology and Evolution. doi:https://doi.org/10.1093/molbev/msx075. Link to the pdf
Abstract
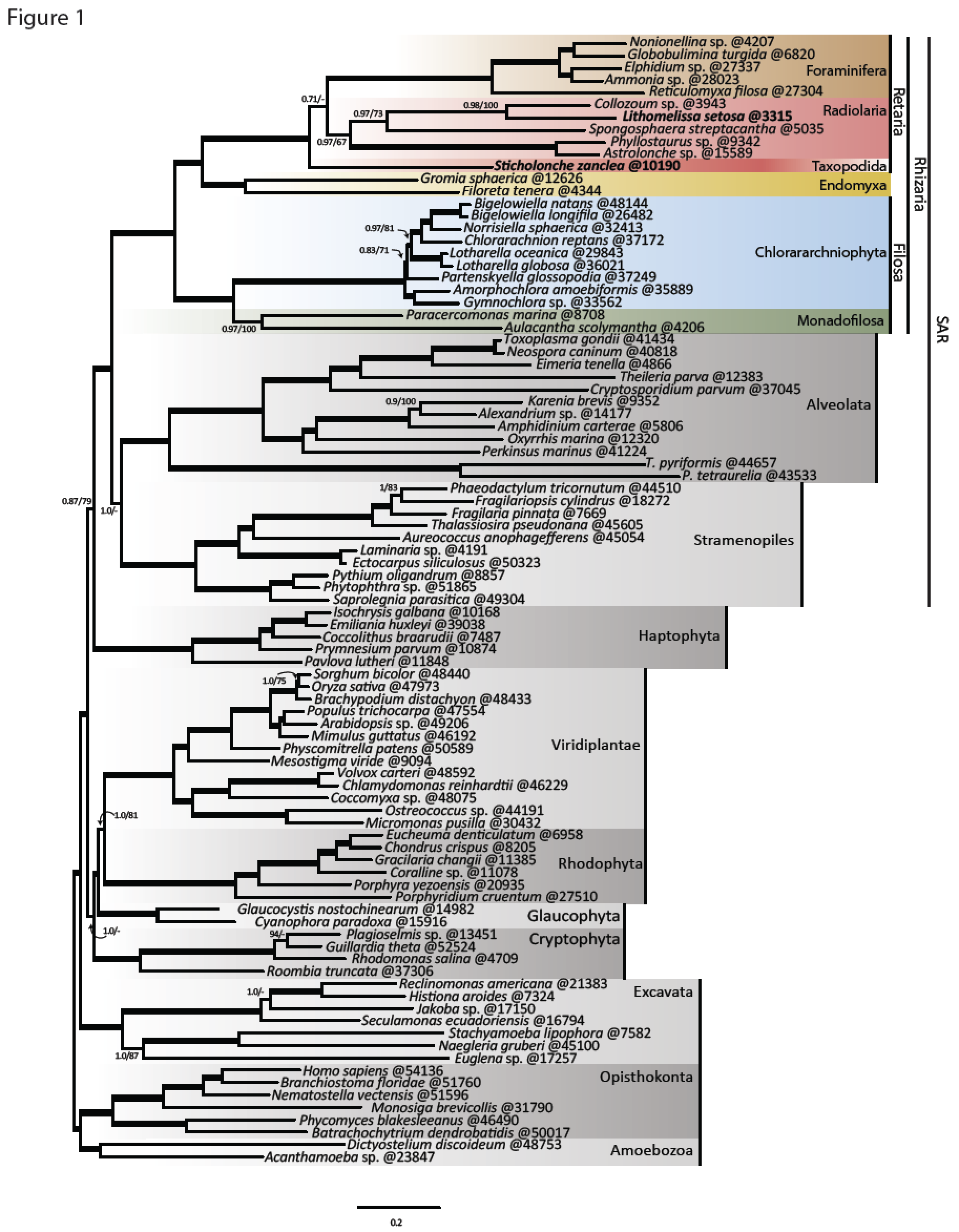
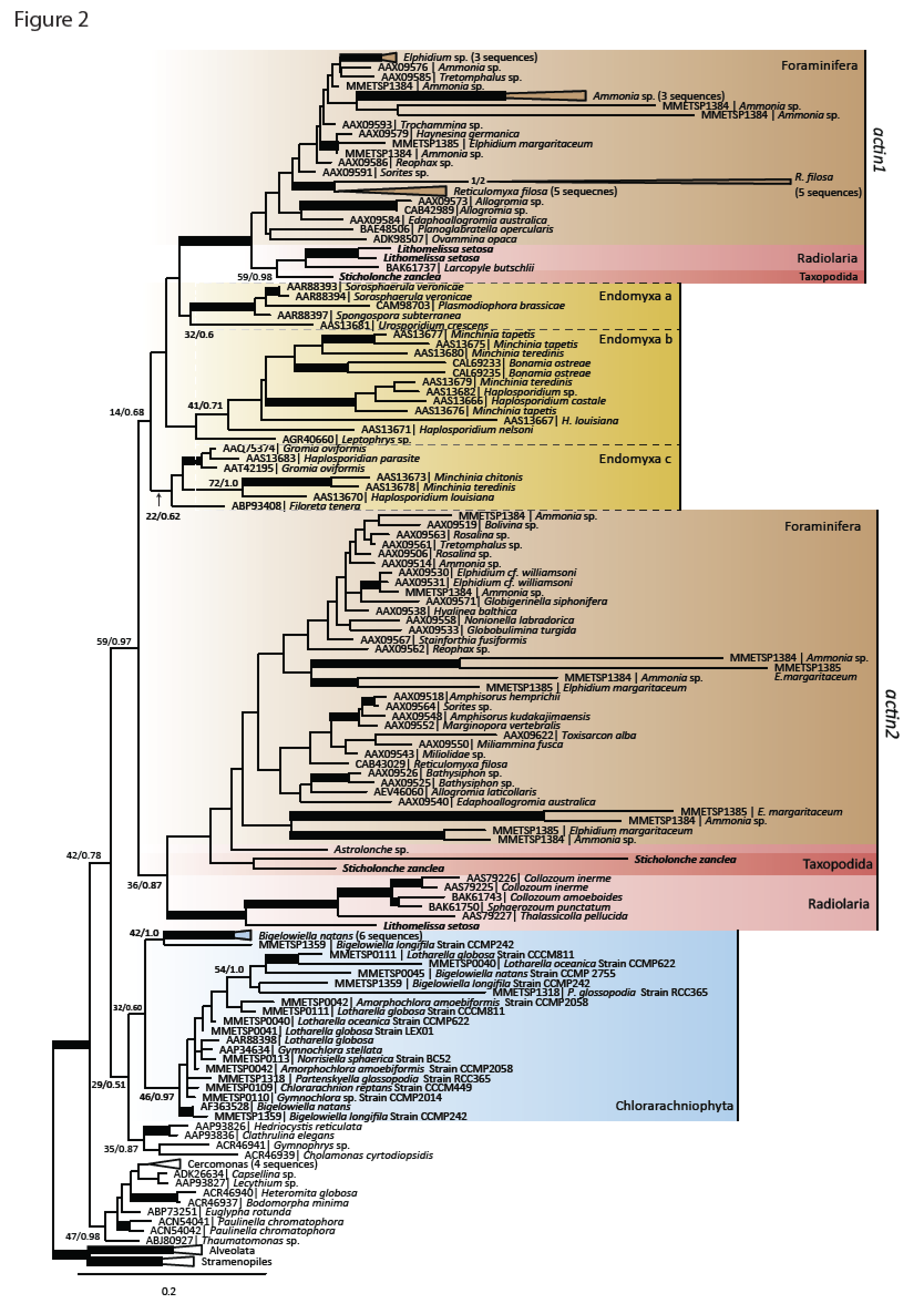
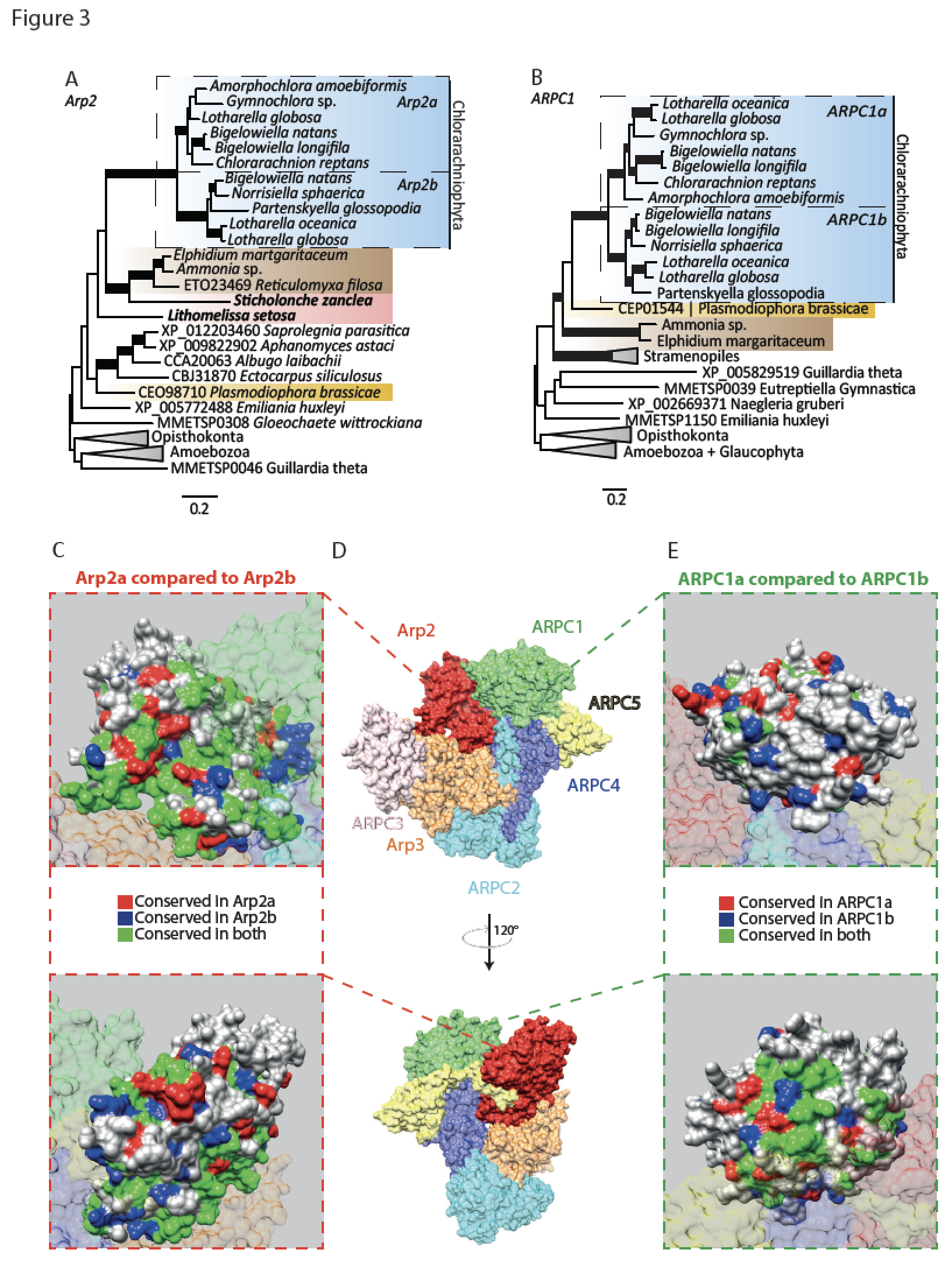
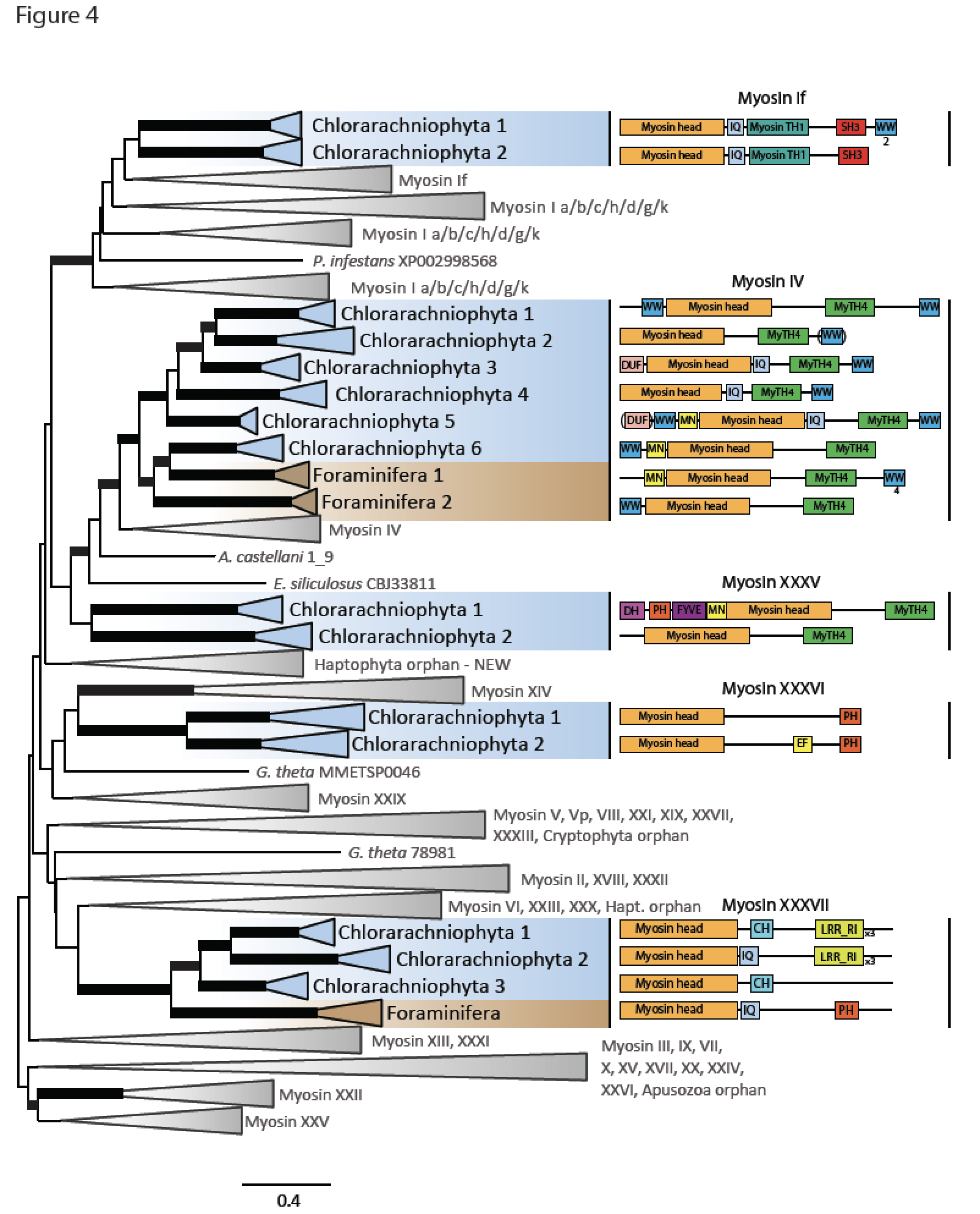
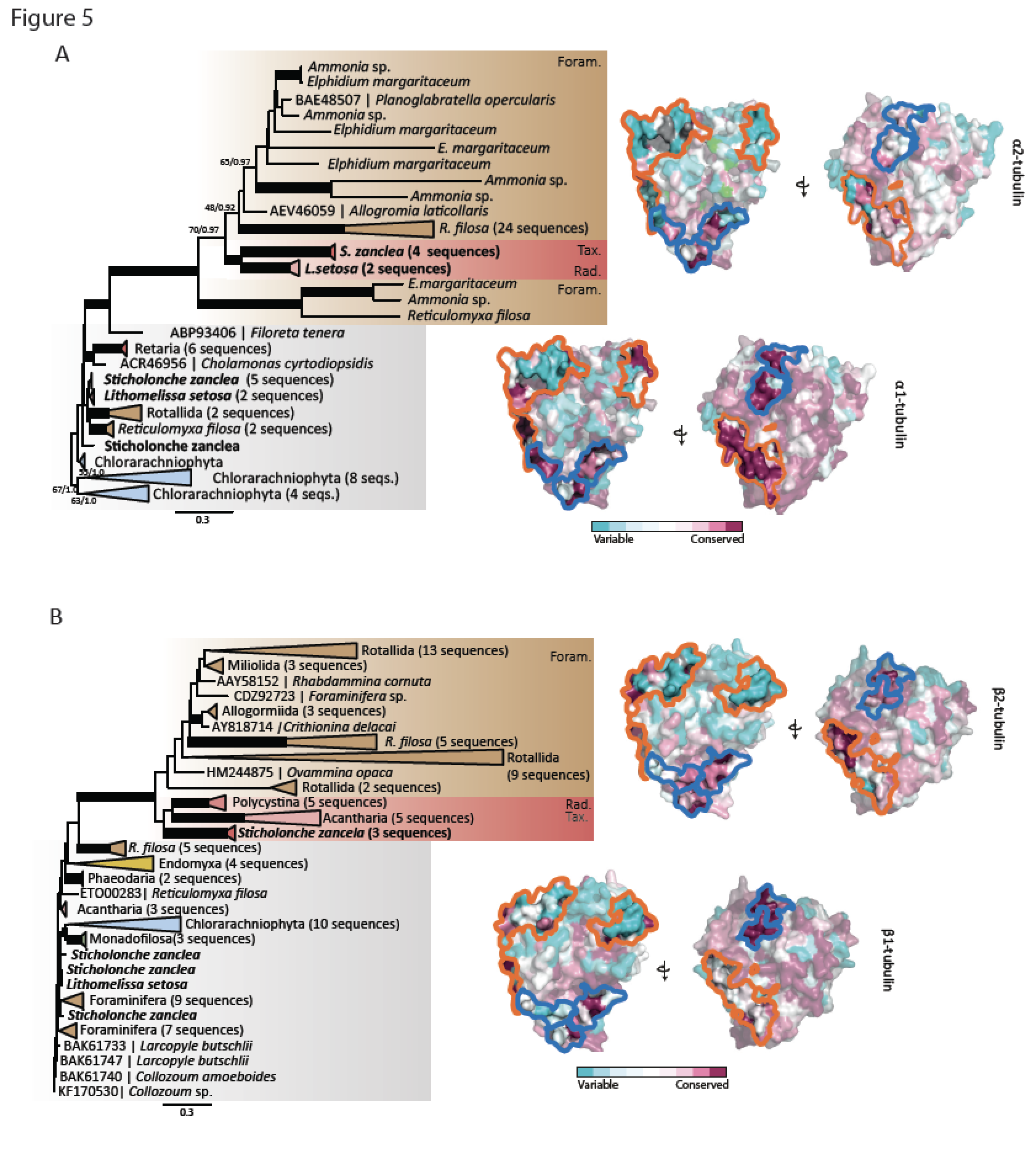
The innovation of the eukaryote cytoskeleton enabled phagocytosis, intracellular transport and cytokinesis, and is responsible for the diversity of eukaryotic morphologies. Still, the relationship between phenotypic innovations in the cytoskeleton and their underlying genotype is poorly understood. To explore the genetic mechanism of morphological evolution of the eukaryotic cytoskeleton we provide the first single cell transcriptomes from uncultured, free-living unicellular eukaryotes: the polycystine radiolarian Lithomelissa setosa (Nassellaria) and Sticholonche zanclea (Taxopodida). A phylogenomic approach using 255 genes finds Radiolaria and Foraminifera as separate monophyletic groups (together as Retaria), while Cercozoa is shown to be paraphyletic. Analysis of the genetic components of the cytoskeleton and mapping of the evolution of these to the revised phylogeny of Rhizaria reveal lineage-specific gene duplications and neofunctionalization of α and β tubulin in Retaria, actin in Retaria and Endomyxa, and Arp2/3 complex genes in Chlorarachniophyta. We show how genetic innovations have shaped cytoskeletal structures in Rhizaria, and how single cell transcriptomics can be applied for resolving deep phylogenies and studying gene evolution in uncultured protist species.