Regulatory RNA at the root of animals: dynamic expression of developmental lincRNAs in the calcisponge Sycon ciliatum
Bråte, J., Adamski, M., Neumann, S. R., Shalchian-Tabrizi, K. and Adamska, M. 2015. Proc. Royal. Soc. B. doi:10.1098/rspb.2015.1746
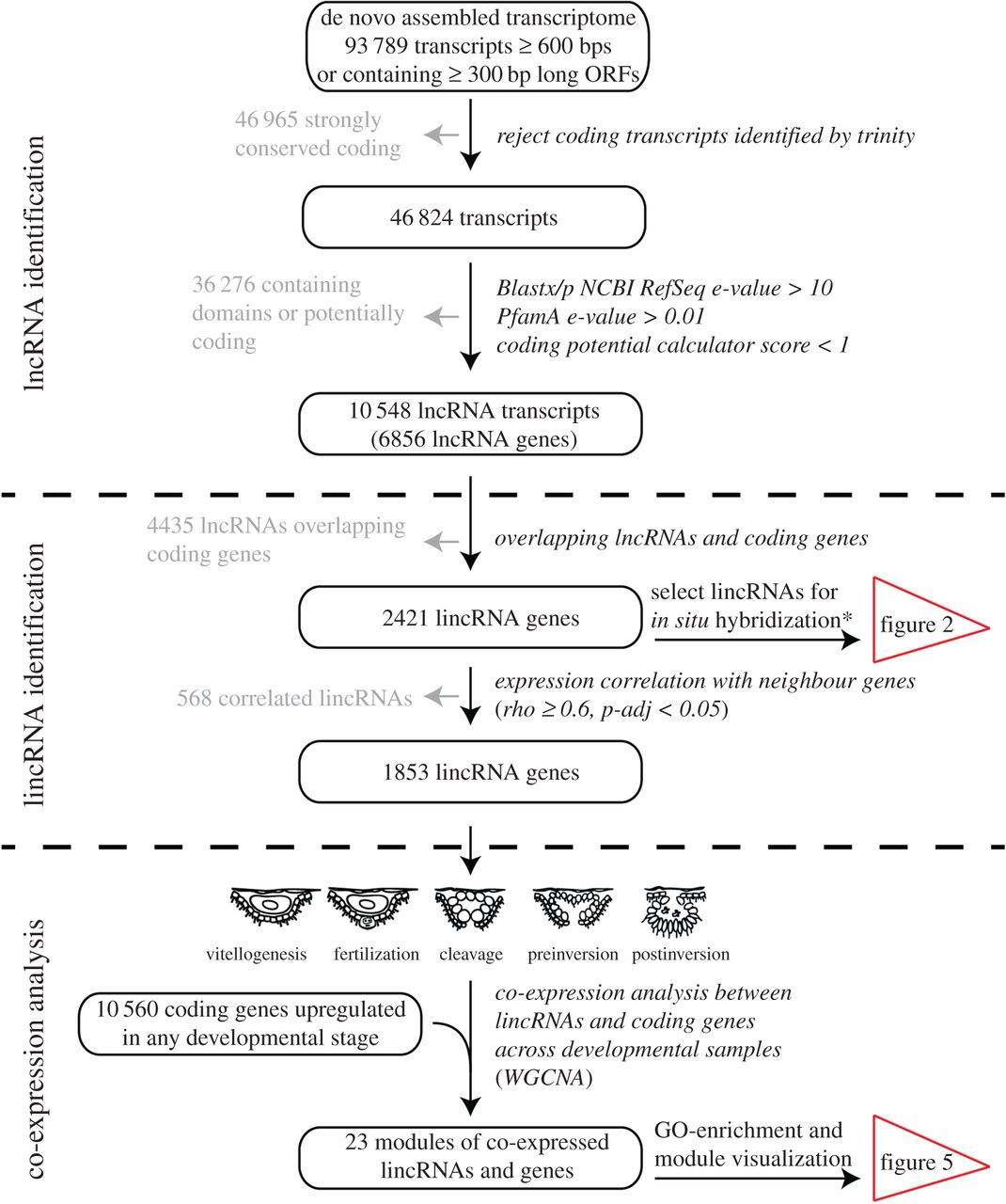
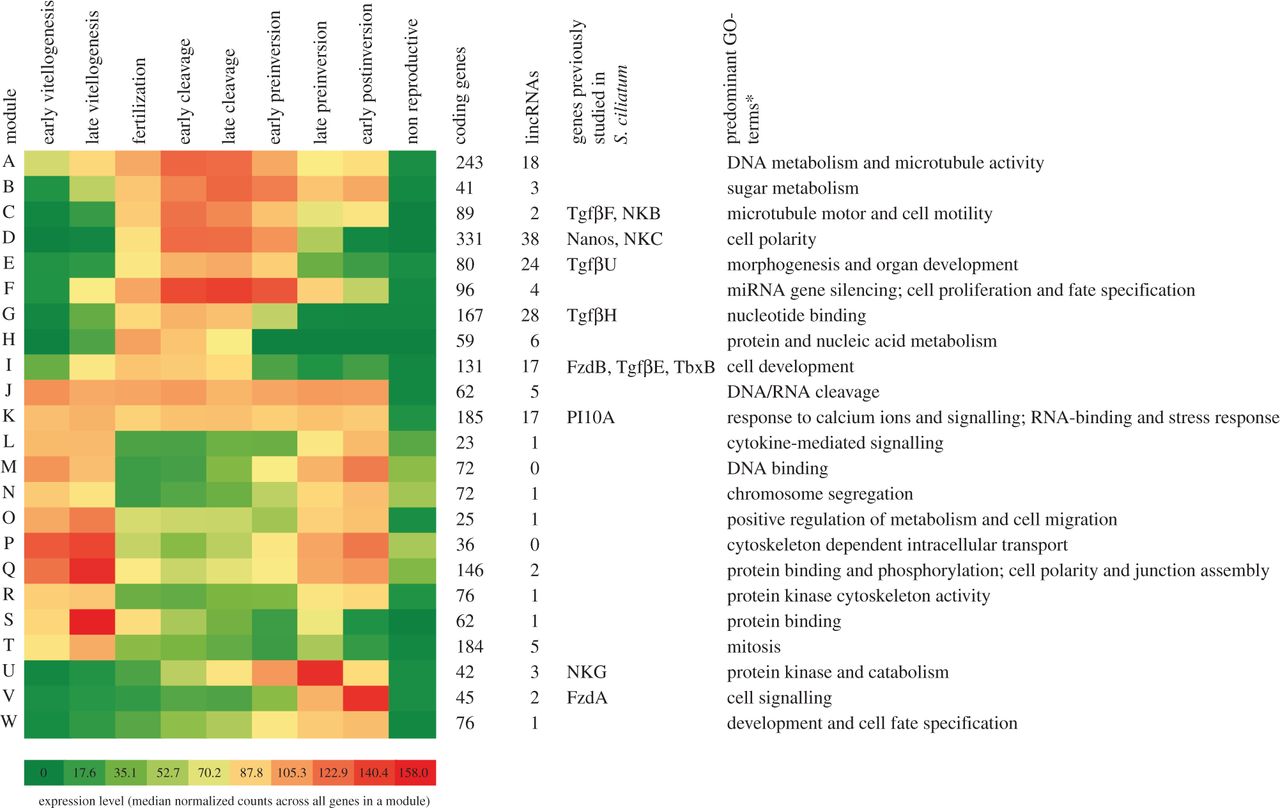
Here we identified and studied the expression of long intergenic non-coding RNAs (lincRNAs) during embryogenesis of the calcareous sponge Sycon ciliatum. This was one of the first studies to use in situ hybridization to visualize the expression of lincRNAs.

Link to the paper on Kudos
Abstract
Long non-coding RNAs (lncRNAs) play important regulatory roles during animal development, and it has been hypothesized that an RNA-based gene regulation was important for the evolution of developmental complexity in animals. However, most studies of lncRNA gene regulation have been performed using model animal species, and very little is known about this type of gene regulation in non-bilaterians. We have therefore analysed RNA-Seq data derived from a comprehensive set of embryogenesis stages in the calcareous sponge Sycon ciliatum and identified hundreds of developmentally expressed intergenic lncRNAs (lincRNAs) in this species. In situ hybridization of selected lincRNAs revealed dynamic spatial and temporal expression during embryonic development. More than 600 lincRNAs constitute integral parts of differentially expressed gene modules, which also contain known developmental regulatory genes, e.g. transcription factors and signalling molecules. This study provides insights into the non-coding gene repertoire of one of the earliest evolved animal lineages, and suggests that RNA-based gene regulation was probably present in the last common ancestor of animals.