Draft genome assembly and transcriptome sequencing of the golden algae Hydrurus foetidus (Chrysophyceae)
Bråte, J., Fuss, J., Jakobsen. K. S. and Klaveness, D. 2019. F1000Research. doi:10.12688/f1000research.16734.1

Abstract
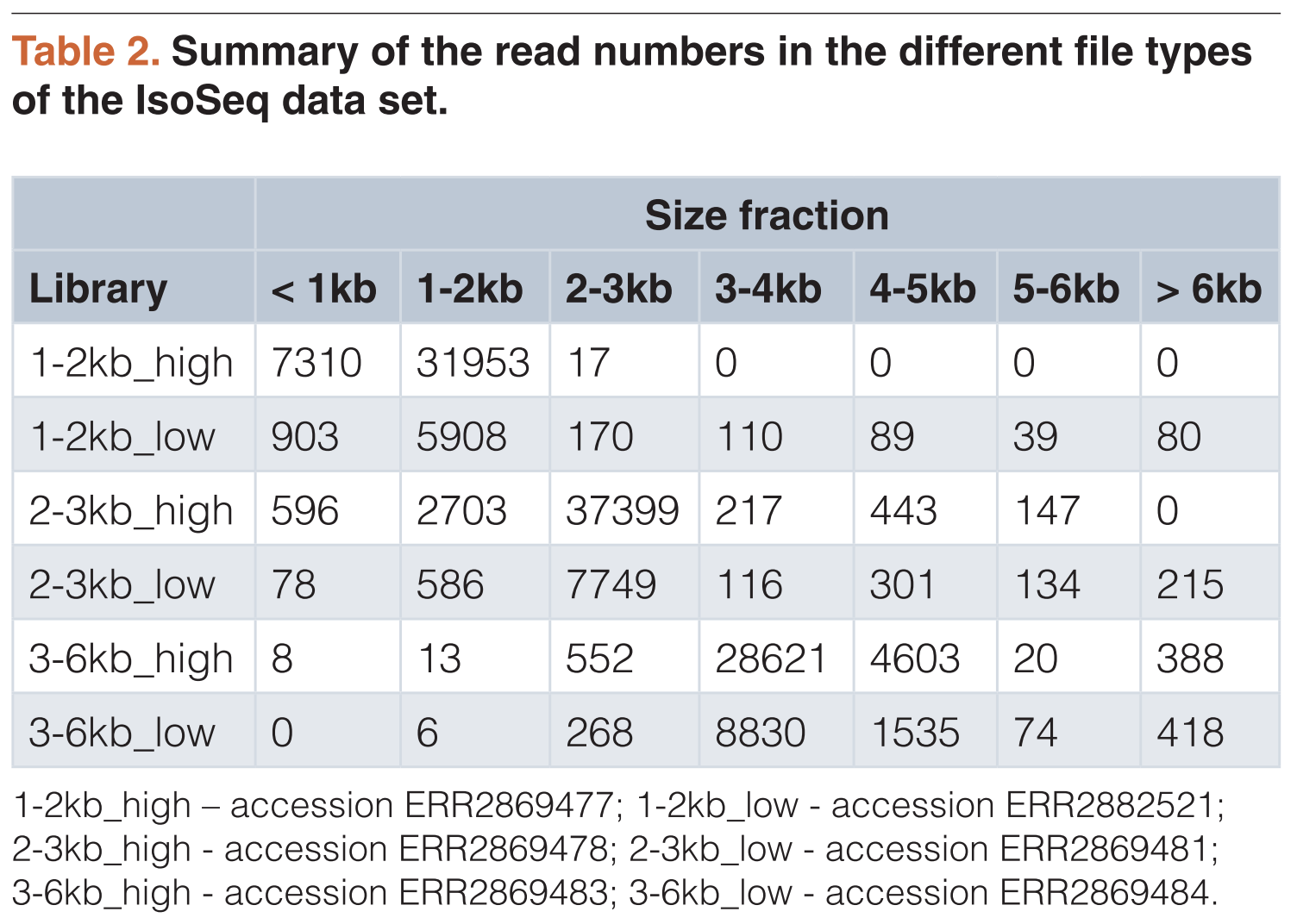
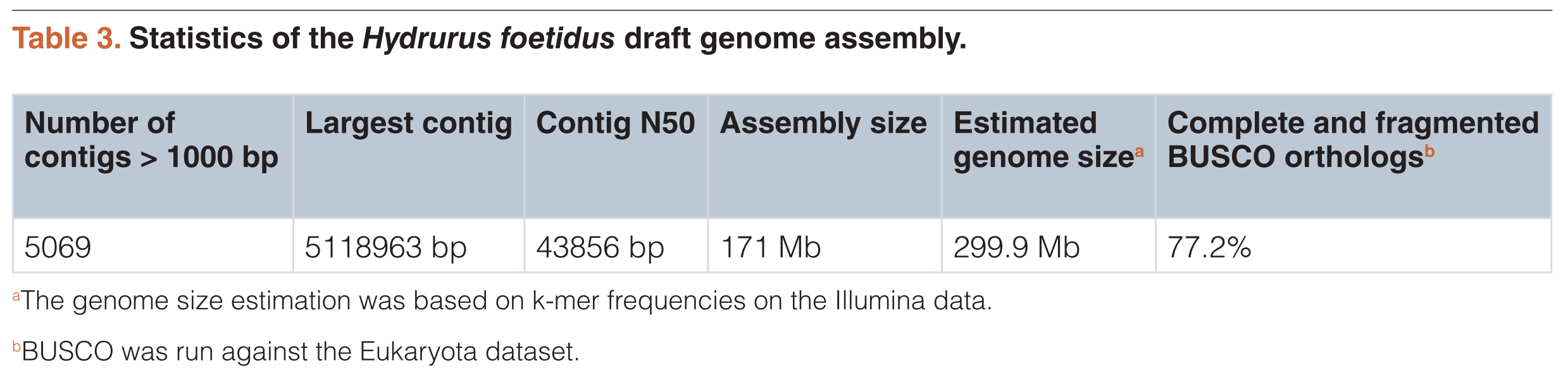
Hydrurus foetidus is a freshwater alga belonging to the phylum Heterokonta. It thrives in cold rivers in polar and high alpine regions. It has several morphological traits reminiscent of single-celled eukaryotes, but can also form macroscopic thalli. Despite its ability to produce polyunsaturated fatty acids, its life under cold conditions and its variable morphology, very little is known about its genome and transcriptome. Here, we present an extensive set of next-generation sequencing data, including genomic short reads from Illumina sequencing and long reads from Nanopore sequencing, as well as full length cDNAs from PacBio IsoSeq sequencing and a small RNA dataset (smaller than 200 bp) sequenced with Illumina. We combined this data with, to our knowledge, the first draft genome assembly of a chrysophyte algae. The assembly consists of 5069 contigs to a total assembly size of 171 Mb and a 77% BUSCO completeness. The new data generated here may contribute to a better understanding of the evolution and ecological roles of chrysophyte algae, as well as to resolve the branching patterns within the Heterokonta.